A Review of Computational Phantoms for Quality Assurance in Radiology and Radiotherapy in the Deep-Learning Era
Article information
Abstract
The exciting advancement related to the “modeling of digital human” in terms of a computational phantom for radiation dose calculations has to do with the latest hype related to deep learning. The advent of deep learning or artificial intelligence (AI) technology involving convolutional neural networks has brought an unprecedented level of innovation to the field of organ segmentation. In addition, graphics processing units (GPUs) are utilized as boosters for both real-time Monte Carlo simulations and AI-based image segmentation applications. These advancements provide the feasibility of creating three-dimensional (3D) geometric details of the human anatomy from tomographic imaging and performing Monte Carlo radiation transport simulations using increasingly fast and inexpensive computers. This review first introduces the history of three types of computational human phantoms: stylized medical internal radiation dosimetry (MIRD) phantoms, voxelized tomographic phantoms, and boundary representation (BREP) deformable phantoms. Then, the development of a person-specific phantom is demonstrated by introducing AI-based organ autosegmentation technology. Next, a new development in GPU-based Monte Carlo radiation dose calculations is introduced. Examples of applying computational phantoms and a new Monte Carlo code named ARCHER (Accelerated Radiation-transport Computations in Heterogeneous EnviRonments) to problems in radiation protection, imaging, and radiotherapy are presented from research projects performed by students at the Rensselaer Polytechnic Institute (RPI) and University of Science and Technology of China (USTC). Finally, this review discusses challenges and future research opportunities. We found that, owing to the latest computer hardware and AI technology, computational human body models are moving closer to real human anatomy structures for accurate radiation dose calculations.
Introduction
The so-called “deep learning,” cashing in on long-time research in the field of machine learning, is expectedly affecting medical imaging and radiological sciences. The “modeling of digital human” in terms of a computational phantom for radiation dose calculations has experienced a large increase in activity in the past two decades, perhaps aided by the latest hype related to deep learning. Radiation dosimetry involves the determination of the amount and distribution pattern of ionizing energy deposited in a material. It is challenging to achieve accurate radiation dosimetry for the following reasons [1]: (1) exposure scenarios are diverse and often include a complex and unique geometrical relationship between the source and the human body; (2) an exposure can involve multiple radiation types, and different types of radiation transverse the human body and interact with tissues according to different radiation physics principles; and (3) the human body consists of a very large number of anatomical structures that are heterogeneous in density and composition. The last point emphasizes the importance of anatomical models in radiation dosimetry because the dose inside a living person is difficult to measure directly. Instead, computational or physical phantoms must be used to estimate the dose delivered to a worker or patient exposed to ionizing radiation. Compared to using physical phantoms, computational approaches are generally advantageous with respect to versatility, efficiency, precision, and safety. Furthermore, internally distributed radiation sources are best handled by computational approaches.
For the past 50 years, researchers have been extremely interested in assessing radiation dosage using computational phantoms in the fields of radiation protection, medical imaging, and radiotherapy. Health physicists often need to estimate the level of internal and external radiation exposure of workers as part of the radiation safety program mandated by federal and state regulations. In diagnostic radiology and nuclear medicine, the imaging process involving energetic X-ray and gamma-ray photons must be optimized to achieve the necessary level of image quality while minimizing potentially harmful radiobiological effects. Radiation therapy aims to deposit lethal doses of radiation to a tumor while preventing the toxicity of healthy tissues and avoiding the risk of developing secondary cancer. While the radiological physics principles have not changed in the past 60 years, computer technologies, especially artificial intelligence (AI) and graphics processing unit (GPU) technologies, have advanced exponentially, leading to a large unforeseen increase in research activities in the field of anatomical modeling for radiation dosimetry in the past two decades.
Observations and Discussion
1. Phantoms
Due to the advantages mentioned above, computational phantoms have been extensively developed by researchers. Eckerman et al. [2] identified a total of 121 computational phantoms that were developed for ionizing and nonionizing radiation dosimetry. Prior to the 1990s, there were fewer than 25 such computational phantoms, but this number surprisingly increased to more than 400 [1]. Today, the number of computational phantoms may even exceed several thousand. In retrospect, the trend of computational phantom development in the past 60 years has clearly mirrored the exponential growth pattern of computer technologies.
In a computational phantom, the explicit definition of the surfaces of an organ in which radiation interactions and energy deposition occur is provided. This definition is similar to the solid-geometry modeling methods widely used in computer-aided design (CAD). Two general modeling methods are of interest: (1) constructive solid geometry (CSG) and (2) boundary representation (BREP) [3–5].
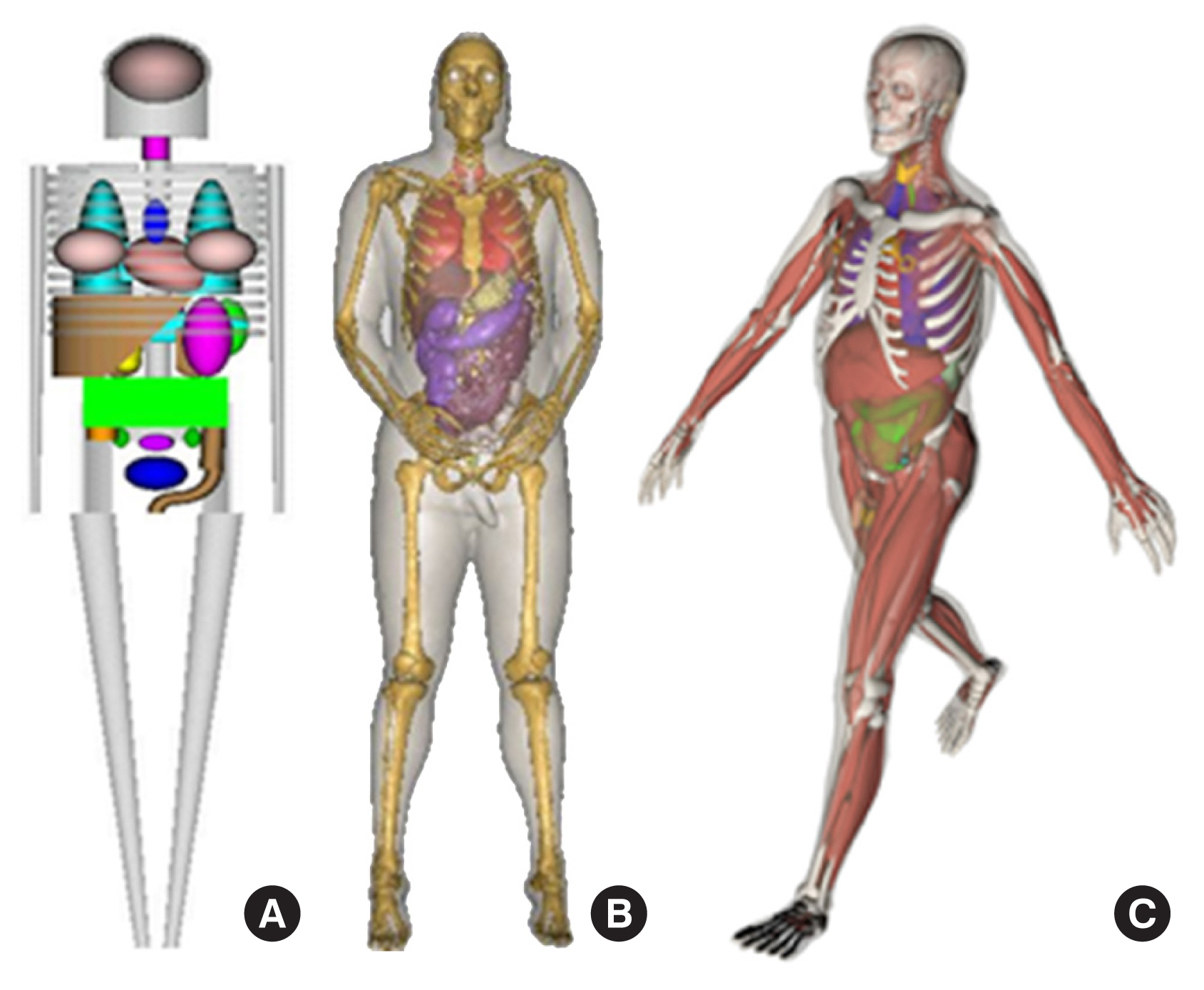
Over the years, literature reviews have been published that focus on a certain period or a particular type of phantom [2, 6, 7]. To date, the most comprehensive literature review was a book edited by Xu and Eckerman [8] that contained contributions by more than 60 authors. More recent information on BREP phantoms was provided in a review by Xu [1]. This review was the first to analyze the trend in the development of approximately 400 computational phantoms over the past 50 years. Based on chronological and technical information in these literature reviews, computational phantoms can be classified into three generations in terms of when they were introduced: (1) stylized phantoms that are based on quadric equations (from the 1960s to the 1980s); (2) voxel phantoms that are based on tomographic images (from the 1980s to present); and (3) BREP phantoms that are based on advanced primitives and are deformable (from the 2000s to present). Fig. 1 compares these generations of computational phantoms, which have different levels of geometric sophistication and anatomical realism.
1) Stylized phantoms (from the 1960s to the 1980s)
The first-generation anthropomorphic computational phantoms were developed to better assess organ doses from internally deposited radioactive materials for workers and patients [2].
In 1959, the International Commission on Radiological Protection (ICRP) used very simple models for the internal dosimetry calculations associated with the Report of ICRP Committee II [9]. In this approach, the total body was represented as a sphere with a 30-cm radius. With the gradual increase in computing power from the late 1950s through the 1960s, the first generation of stylized anthropomorphic phantoms was developed at Oak Ridge National Laboratory (ORNL) [10]. Using CSG modeling techniques involving shapes such as elliptical cylinders and cones, the Fisher-Snyder adult phantom was developed.
In 1969, Snyder and his colleagues [11] reported the first heterogeneous phantom that became known as the “MIRD-5 phantom,” a name derived from the Medical Internal Radiation Dosimetry (MIRD) Committee of the Society of Nuclear Medicine (SNM) that adopted the phantom. This phantom consisted of a skeleton, a pair of lungs, and soft tissue. The representation of the internal organs is roughly based on the mathematical model because simple equations capture only the most general description of the position and geometry of each organ. The original model was designed to represent a healthy “average” adult male, which was referred to as Reference Man by the ICRP. The characteristics of Reference Man were the result of an extensive review of medical and other scientific literature on the European and North American populations [12]. Reference Man was defined as a 20- to 30-year-old Caucasian male, 70 kg in weight and 170 cm in height; the height of Reference Man was later changed from 170 cm to 174 cm. In 1978, Snyder [13] published a detailed set of specific absorbed fractions using an improved version of their heterogeneous phantom, which contained more than 20 organs and more detailed anatomical features.
Significant efforts were undertaken at ORNL during the mid-1970s to develop individual pediatric phantoms. Hwang et al. [14] designed three “individual phantoms”: a neonatal phantom, a 1-year-old phantom, and a 5-year-old phantom. A separate effort was undertaken by Jones et al. [15] to develop a 15-year-old phantom, and Deus and Poston [16] undertook the design of a 10-year-old phantom after completing the other four designs. Based on previous work, Cristy [17] reported the development of a new series of stylized phantoms in 1980 and then collaborated with Eckerman in 1987 on the report ORNL/TM-8381 [18]. The “family” of this series of phantoms included an adult male, a newborn, and individuals of 1, 5, 10, and 15 years of age; the 15-year-old phantom also represents an adult female when additional anatomical features are added. As shown in Fig. 2, each phantom consists of three main parts: (1) an elliptical cylinder representing the trunk and arms; (2) two truncated circular cones representing the legs and feet; and (3) an elliptical cylinder that sits on top of the cylinder, and a half ellipsoid that sits at the upper end of the elliptical cylinder, which represents the head and neck, respectively. Attached to the legs is a small area with a flat front surface to contain the testes. The female phantom included two ellipsoids attached to the torso to represent breasts (not shown in Fig. 2). The arms were embedded in the torso. Minor appendages, such as the fingers, feet, chin, and nose, were omitted.
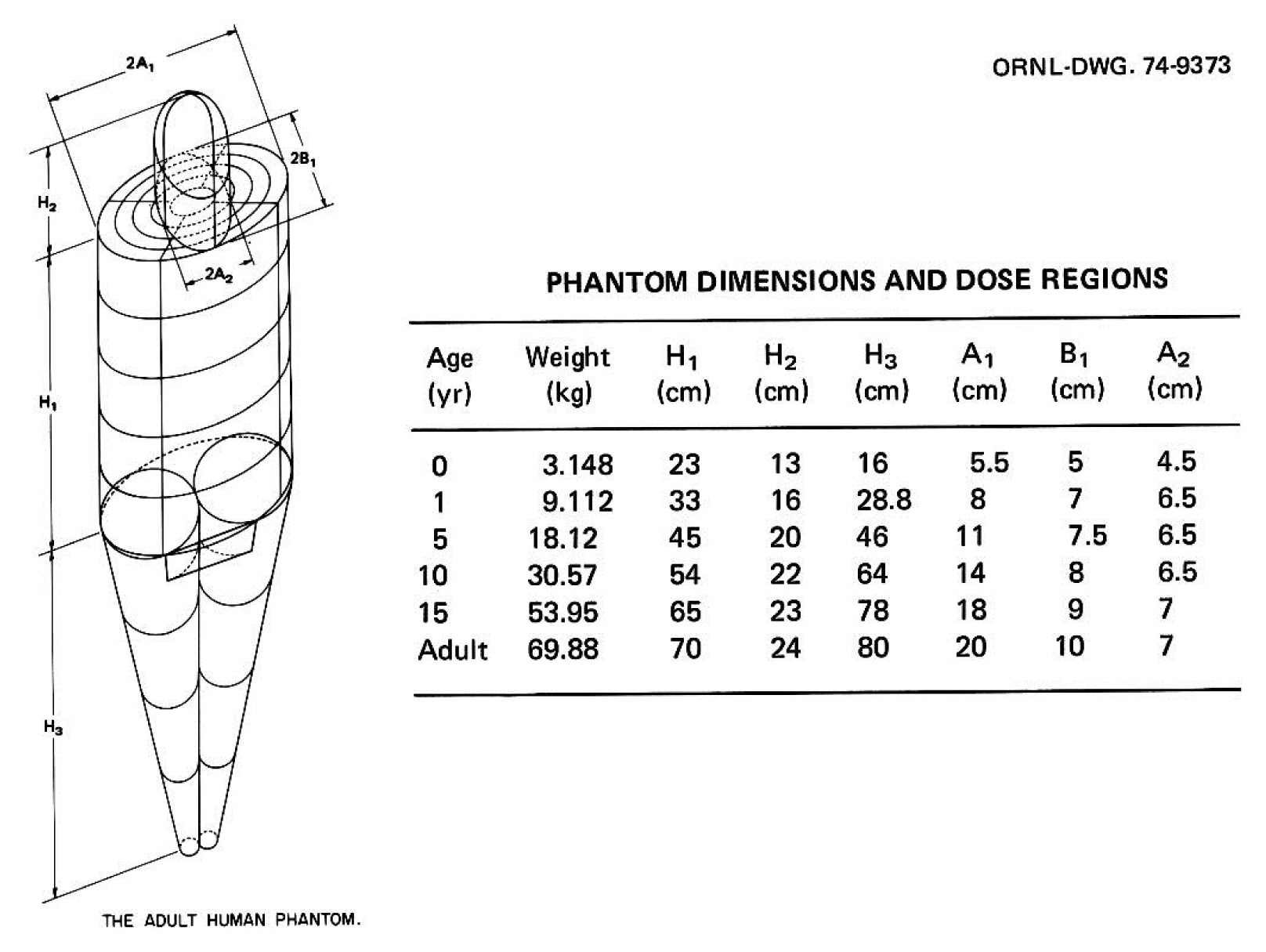
The adult male phantom and its dimensions. Similar descriptions and diagrams were purposely followed in a series of ORNL technical reports by Snyder [13], Cristy [17], and Cristy and Eckerman [18]. Adapted from Xu [1]. Phys Med Biol. 2014;59(18):R233–R2302 with the permission of IOP Publishing. ORNL, Oak Ridge National Laboratory.
In 1995, Stabin and his colleagues [19] at ORNL adapted the phantom of an adult female in this family to represent a pregnant woman in the third trimester. Three sets of stylized pregnant female phantoms were used in various internal nuclear medicine applications.
At the same time that ORNL’s Cristy and Eckerman [18] were working to improve the MIRD-5 phantom, a group at the National Research Center for Environment and Health (GSF) in Germany, which is now known as the German Research Center for Environmental Health (HZM), used the anatomical descriptions of the hermaphrodite MIRD-5 phantom to develop a pair of gender-specific adult phantoms, which were later known as ADAM and EVA, for external dosimetry studies. The EVA phantom was based on the ICRP reference organ quality analysis. All relevant volumes of the MIRD-5 phantom were scaled down, and the overall mass ratio was 0.83. Then, the female organ masses were transformed to create space for neighboring organs. Finally, sex-specific organs, such as the testes for ADAM and the ovaries, uterus, and breasts for EVA, were implanted. The chin was used to create a more realistic external irradiation geometry for the thyroid gland by removing part of the neck. The female breasts were connected to the torso of the EVA phantom by two oval sections. There were some subtle differences in the anatomy, such as breast size, between EVA and the phantom reported by Cristy and Eckerman [18].
During roughly the same period of time, the computational anatomical man (CAM) and computational anatomical female (CAF) phantoms were developed by the National Aeronautics and Space Administration (NASA) and consist of 1,100 unique geometric surfaces and 2,450 solid regions [20]. CSG modeling technology was used to explicitly model the internal geometric structures of the human body (such as organs, cavities, bones, and bone marrow). An analytical computer code (not based on Monte Carlo methods) called CAMERA was also developed for performing dose calculations with the CAM and CAF phantoms. Unfortunately, detailed information on the CAM and CAF phantoms was not publicly available until the early 2000s [21]. It is interesting to note one unique exterior anatomical feature of these phantoms: unlike the MIRD-5 phantom, the arms were separated from the trunk. Successors to these phantoms were developed almost simultaneously [1].
Since the publication of the stylized dosimetry model of Snyder [13] in the MIRD Pamphlet 5 Revised, the MIRD Committee of SNM refined several internal organs to support the development of radiopharmaceutical tracers and therapeutic nuclear medicine. Modifications to the MIRD stylized model were published as MIRD Pamphlets, which include equations for new geometries, tables for absorbed fractions of energy for monoenergetic photons and electrons, and tables for radionuclide S-values. In 1999, the MIRD Committee adopted six new head and brain models for the following phantoms: newborn, 1-year-old, 5-year-old, 10-year-old, 15-year-old (which also represents the average adult female), and adult male phantoms [22]. Similar to previous stylized models, simplified geometric shapes were used to represent the different regions of the head and brain; volumes were derived from published reference masses and shapes from the analysis of magnetic resonance imaging (MRI) images. Later, the MIRD Committee also adopted a series of age-dependent stylized kidney models. These models have been widely used in therapeutic nuclear medicine for the prediction of renal toxicity [23].
The stylized modeling technique was also adopted for medical imaging applications. For example, the mathematical cardiac torso (MCAT) phantom was developed by a research group led by Benjamin Tsui (who is currently with Johns Hopkins University) at the University of North Carolina for use in single-photon emission computed tomography (SPECT) and positron emission tomography (PET) [24–26]. Segars, a member of the group from Johns Hopkins University, later developed more advanced phantoms.
For radiation dosimetry studies, Xu [1] also summarized other stylized phantoms, including an embryo phantom and a fetus phantom [27], a Korean adult phantom [28], a Japanese 9-month-old phantom [29], a Chinese mathematical phantom (CMP) [30], a standard Korean male phantom [31], a new mathematical model to simulate the reference male bottle mannikin absorber (BOMAB) phantom [32, 33], and a converted MIRD-type mathematical phantom in NURBS (non-uniform rational B-splines)/voxels [34].
For nearly 50 years after the first stylized phantom was reported, these anatomically simplified phantoms were used as the de facto “standard” representations of the ICRP Reference Man methodology, which is based on “population-average” 50th-percentile anatomical parameters specified in ICRP Publication 23 [12] and ICRP Publication 89 [35]. Applications of stylized phantoms ultimately included many aspects of radiation protection, radionuclide therapy, and medical imaging [36]. In addition, national and international agencies have adopted organ dose estimates based on these stylized phantoms in guidelines and regulations related to industrial and medical uses of ionizing radiation. For computed tomography (CT) dose reporting, commercial software systems developed before the 2000s were based on stylized patient models, which are known to cause large errors when low-energy X-rays are utilized [37]. In epidemiological studies, e.g., Stovall et al. [38], stylized models were used to derive dose-response relationships between Japanese atomic bomb survivors and radiotherapy patients. In the early 1990s, Xu and his colleagues [39–42] used stylized phantoms, 24 PCs with Intel 486 processors, and MCNP code (version 3) to calculate organ doses and personnel dosimeter responses for nuclear power plant workers.
Although stylized phantoms made it possible to perform Monte Carlo computations during a time when computers were much less powerful, the original developers realized the obvious shortcomings of these phantoms. The human anatomy is too complicated to be realistically modeled with a limited set of surface equations. Many anatomical details in these models were missing, which sometimes led to inaccurate dosimetric results [8].
2) Voxel Phantoms (from the 1980s to present)
The development of anatomically realistic models became feasible in the 1980s with the development of powerful computer and tomographic imaging technologies such as CT and MRI. Xu [1] summarized the construction of a total of 84 voxel phantoms, usually from three types of tomographic images: CT and magnetic resonance (MR) images of live subjects and cross-sectional photographs of cadavers. In two previously published review articles, Caon [6] reported a total of 21 voxel phantoms, and Zaidi and Xu [7] reported 38 voxel phantoms.
In terms of the developmental process, tomographic (voxel) phantoms are fundamentally different from stylized phantoms. A tomographic image dataset consists of many slices, and each slice displays an anatomical two-dimensional (2D) pixel map. The three-dimensional (3D) volume of a voxel is measured by multiplying the pixel size by the thickness of an image slice. Unlike stylized phantoms based on quadric surface equations, a voxel phantom contains a large number of small cubes grouped to represent various anatomical structures. However, both quadric surface equations and cubic voxels belong to the same type of CSG geometries.
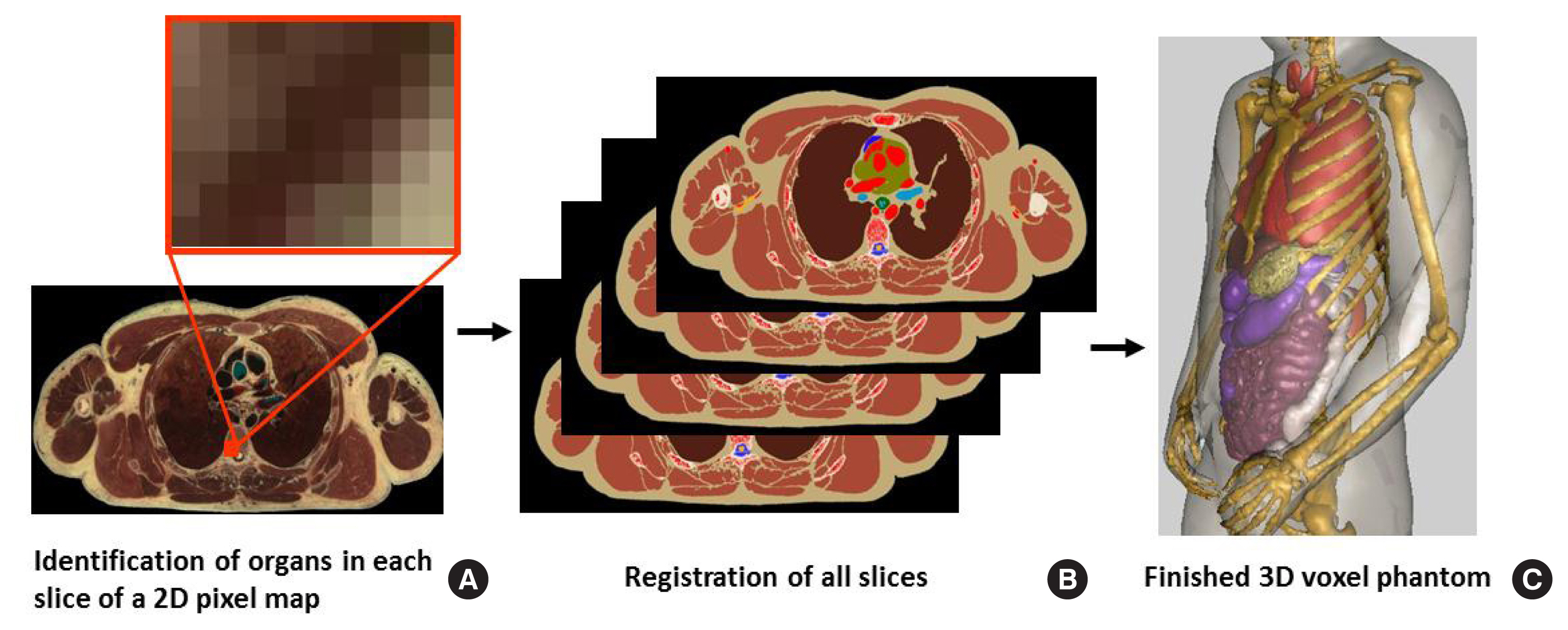
Generating a tomographic (voxel) phantom generally includes four steps. (1) Obtain a set of tomographic images (e.g., CT, MR, or anatomical photography) covering the entire volume of the body. (2) Identify (or segment) organs or tissues of interest (e.g., lungs, liver, or skin) from the original image slice by assigning every pixel an identification number. (3) Specify the density (e.g., soft tissue, hard bone, or air) and chemical composition of organs and tissues. And (4) register the segmented image slices into a 3D volume that can be used for 3D visualization (for checking anatomical structures) and for Monte Carlo calculations. Fig. 3 illustrates these steps using the visible photographic man (VIP-Man) phantom [43].
The rapid development of voxelized phantoms by researchers from different countries was discussed in detail by Xu [1]. Gibbs of Vanderbilt University and his colleagues [44–46] were the first to work on creating image-based radiation dose phantoms. However, Zankl and her colleagues [47–52] from German, who are one of the most well-known groups, started to use CT imaging in the late 1980s for what eventually became a family of 12 voxel phantoms: BABY, CHILD, DONNA, FRANK, HELGA, IRENE, GOLEM, GODWIN, VISIBLE HUMAN, LAURA, KLARA, and KATJA. Recognizing the potential of voxel phantoms, the ICRP [53] stated that “An important issue for Committee 2 is the substitution of an anatomically realistic voxel phantom, obtained digitally in magnetic resonance tomography and/or computed tomography, for the MIRD phantom which is a mathematical representation of a human body.” The ICRP Committee 2 has a dose calculations (DOCAL) task group, which is directly responsible for developing sets of standard voxel phantoms. Later, the team led by Zankl [54] made major modifications to the GOLEM and LAURA phantoms, resulting in new phantoms called REX and REGINA, which were released to the public as the ICRP adult Reference Male and Reference Female [55].
Zubal et al. [56] from Yale University in the United States published a head-torso model called VoxelMan, which was developed from CT images. Other researchers revised the original VoxelMan data, which was publicly available, to develop the MANTISSUE3-6 and VOXTISS8 [57, 58], MAX (Male Adult voXel) [59], FAX (Female Adult voXel) [60], MAX06, and FAX06 phantoms [61]. The work by Kramer et al. [61] was one of the earliest efforts to create ICRP-89 compatible voxel phantoms for radiation protection dosimetry. In 2008, Akkurt et al. [62] from ORNL reported their work involving a mixture of voxel and stylized geometries.
In 1996, Dimbylow [63] from the United Kingdom used MR images for an adult male phantom known as NORMAN to calculate exposure levels to nonionizing electromagnetic fields [64]. Later, this phantom was used to estimate organ doses from external and internal photon sources [65]. In 2005, Dimbylow [66] developed an adult female phantom, NAOMI, from MR scans. In 2005, a revised version of the NORMAN phantom, called NORMAN-5, was created in Italy to derive external photon dose data [67]. One year later, Dimbylow and his colleagues [68, 69] merged NAOMI with stylized and voxelized fetal phantoms to create a series of hybrid phantoms of pregnant women.
In 1999, Caon et al. [70, 71] from Australia reported a torso phantom named ADELAIDE, which was generated based on CT images of a 14-year-old girl. Caon [6] later reviewed his and other researchers’ experiences on voxel phantoms.
Work at the Rensselaer Polytechnic Institute (RPI) on voxel phantoms began with the VIP-Man voxel phantom [43]. The uniqueness of this phantom was that it was developed using cross-sectional color photographic images of an adult male cadaver. These images were from the visible human project (VHP) sponsored by the National Library of Medicine [72]. Ultrafine color image analysis allowed many small and radiosensitive tissues, such as the stomach mucosa, skin, and red bone marrow, to be clearly defined in the phantom. The VIP-Man phantom was used in many studies and will be discussed later in this article. It is worth noting that the CT data from the VHP were also used by Zankl et al. [48] from Germany and Segars [101] from the United States. The RPI group later reported a voxel pregnant female phantom using CT images of a 30-week pregnant patient and compared internal dose data with those derived from a stylized phantom [73, 74].
Xu and his coworkers [43] proposed the concept of a “digital human” by stating that “VIP-Man is digital, and it can be easily adopted for applications beyond radiation transport by coupling with physical properties that are electrical, thermal, chemical, mechanical, or biological. When these become technically possible in the future, the reality of a ‘virtual digital human’ for every citizen in the ‘digital society’ will be within reach.” After 20 years, we are steps closer to realizing that vision.
Realizing the need for more phantoms representing children of different ages, Bolch and colleagues [75–77] from the University of Florida developed a series of voxel phantoms for children, representing children from newborn to 15 years of age. The purpose of the University of Florida pediatric series was to provide a reference library of phantoms that could be matched to the age-specific organ dose assessment of individual patients.
Voxel phantoms were developed and used by groups from many countries. Japanese voxel phantoms include Otoko, Onago JM (Japanese adult Male voxel), JM2, and JF (Japanese adult Female voxel) [78–82], TARO and HANAKO [83], and deformed phantoms representing children of 3, 5, and 7 years of age [84]. Korean voxel phantoms include Korean Man (KORMAN), Korean Typical MAN-1 (KTMAN-1), Korean Typical Man-2 (KTMAN-2), High-Definition Reference Korean (HDRK), and Korean WOMAN (KORWOMAN) [19, 85–87]. Chinese voxel phantoms include Chinese Man (CNMAN) [88], Visible Chinese Human (VCH) [89–91], Chinese Voxel Phantom (CVP) [92, 93], and Reference Taiwanese Adult phantoms [94]. Voxel phantoms by groups from other countries include NUDEL (NUmerical moDEL) from Italy [95], work from France [96, 97], work from Austria [98], work from Iran [99], and work from India [100].
3) BREP phantoms (from the 2000s to present)
As detailed by Xu [1], in the past ten years, more than ten groups around the world have contributed to the astonishing surge in the development of the BREP phantom. As of 2014, a total of 183 BREP-based phantoms have been reported globally. BREP phantoms are still being reported in the literature and will continue to contribute to the exponential growth pattern of phantoms that began 60 years ago.
One of the earliest works was described in a Ph.D. thesis at the University of North Carolina using NURBS-based techniques [101]. The famous NURBS-based cardiac-torso (NCAT) phantom was developed from the Visible Human CT image dataset, and its 3D anatomy was later extended into the 4th dimension (4D) to simulate cardiac and respiratory motions. The beating heart model of the 4D NCAT was based on the 4D labeled MRI data of a real patient. The 4D NCAT phantom provides a very large improvement, including more realistic anatomy and new physiology, over the stylized MCAT phantom, which was also worked on by the same research group [102]. The 4D NCAT phantom has been widely used, especially in nuclear medicine imaging research, to evaluate and improve myocardial SPECT imaging. A 4D digital mouse phantom named MOBY was also developed [102–104]. Later, the group that developed MOBY also reported the extended cardiac torso (XCAT) family, which included 35 males and 23 females 4D computational phantoms [105]. The NCAT and XCAT phantoms have been used by other research groups [106–112].
Xu et al. [113] from RPI reported a set of phantoms at the end of the 3-, 6-, and 9-month gestation periods, called RPI-P3, RPI-P6, and RPI-P9, respectively, using BREP modeling techniques. Later, they reported a pair of standardized phantoms called RPI Adult Male and Adult Female. These phantoms were carefully adjusted to match ICRP-89 reference values for more than 70 organs and 45 bones, including the cortical bone, spongiosa, and cavities, as well as muscles [114]. The Adult Male and Adult Female phantoms from RPI were also expanded into phantoms of specific weights, representing the 5th, 25th, 50th, 75th, and 95th weight percentiles, female phantoms with different breast sizes to study the effect on the lung counting of internally deposited radionuclides [29], and 10 phantoms representing overweight and obese individuals with body mass index (BMI) from 26 to 48 kg/m2 [115]. A unique type of posture-specific phantom was also reported by this group to more realistically model how people behave in real-world radiation environments, including a pair of walking phantoms [116], sitting phantoms [117], and computational human for animated dosimetry (CHAD) phantoms with adjustable postures defined by a motion capture system for critical accidents [118]. Fig. 4 shows the process of using motion capture to create a real sequence of worker actions.
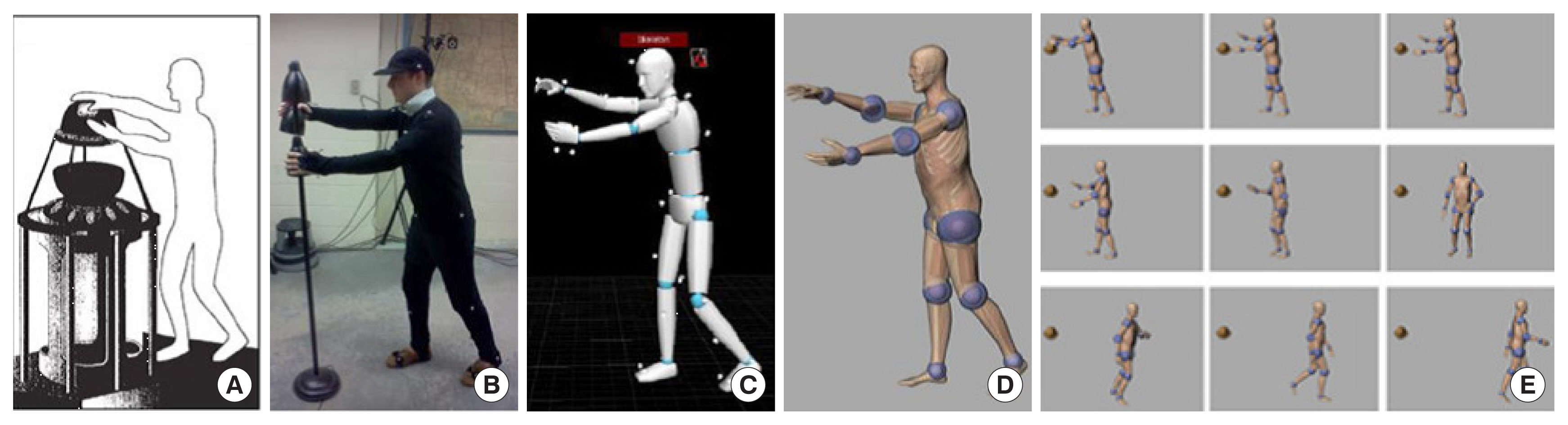
Motion capture technology was used to develop a realistic posture sequence for a critical accident. (A) A worker was exposed to a critical incident and died 66 hours later. (B) An actor reconstructed the postures of the worker using motion capture. (C) The postures of the actor were recorded sequentially. (D) The CHAD phantom recreated the same sequential postures as the actor. (E) A total of nine postures were used for Monte Carlo dose calculations. Adopted from Ref [1] with permission. CHAD, computational human for animated dosimetry.
In a series of papers, the research team at the University of Florida reported their research on “hybrid” NURBS/voxel family phantoms, including male, female, and children of different ages [119–122]. Later, this group also reported a family of NURBS-based fetal phantoms [123]. One important contribution of this group to the literature was in the field of bone marrow dosimetry. For the purpose of radiation protection, photon or neutron dose-response functions could report the active marrow and endosteum dose by tallying photon or neutron fluence in spongiosa regions of the skeleton [124]. Using anatomically realistic phantoms developed at the University of Florida, Johnson et al. [125] reported using the 3-factor method as an alternative to the dose-response function for photon skeletal dose. This method was also applied to neutrons by Bahadori et al. [126]. The University of Florida hybrid adult male phantom has been widely used in dosimetric research [69, 126–132].
Additional BREP phantoms were also mentioned in a review by Xu [1]. A group from Vanderbilt in the United States reported a “family” of adult and pediatric phantoms by adapting the NURBS-based NCAT adult male and female phantoms [133, 134]. A team from Brazil also reported work on a series of BREP phantoms, which included FASH (Female Adult meSH) and MASH (Male Adult meSH) [135, 136]. A team from France developed a series of female torso phantoms for in vivo lung monitoring [137]. Later, a separate project at the Institute for Radiological Protection and Nuclear Safety (IRSN) used the CAESAR database to create a library of 25 whole-body male phantoms [138]. A team from Switzerland reported the Virtual Family, which is a series of BREP-based phantoms used for electromagnetic exposure calculations using MR images [139, 140]. A series of nine phantoms representing a pregnant female in each gestational month was developed at the University of Houston to study the effects of radiofrequencies emitted from various electronic devices [141]. Gu et al. [142] of the Center for Devices and Radiological Health (CDRH) developed a series of high-resolution heart phantoms for accurate dosimetric calculations. Researchers at Hanyang University in Korea used 3D-DOCTOR software to convert the voxel phantom Visible Korean Human-Man (VKH-Man) into a polygon surface phantom and directly implemented this phantom into Geant4 code to circumvent prior limitations [143, 144].
However, the aforementioned BREP phantoms suffer from a critical technical issue, as they are less compatible with existing Monte Carlo radiation transport codes. NURBS phantoms cannot be directly used in Monte Carlo codes [145]. In addition, only a few Monte Carlo codes, such as Geant4, can transport a particle in the polygon surface phantom geometry. Even in this case, the calculation speed is very slow compared to when the voxel geometry is used. Kim et al. [143] showed that for the purposes of photon simulations in Monte Carlo dose calculations, a polygonal surface phantom was approximately 70–150 times slower than a corresponding voxel phantom. Therefore, it is a common practice to convert the surface phantom into the voxel geometry via the “voxelization” process [145, 146], thereby reverting to the inherent limitations of the voxel phantom.
To avoid voxelization of the polygonal mesh (PM), in 2014, Yeom et al. [147] proposed the conversion of the geometrical format of the PM into a tetrahedral mesh (TM) by using a “tetrahedralization” algorithm. The advantage of this approach is that the conversion does not distort the geometry; that is, the converted TM phantom maintains exactly the same organ shapes as in the original PM phantom. In addition, the TM phantom can be directly used in the Monte Carlo simulations. The results showed that in Geant4, the TM phantom is faster than the PM phantom by several orders of magnitude.
Acknowledging both limitations of the voxel geometry and advantages of the TM geometry, the ICRP Task Group 103 converted the voxel-type ICRP-110 reference phantoms into the adult male and adult female TM phantoms [148]. These TM phantoms are called mesh-type reference computational phantoms (MRCPs). They were completed and tested with the Geant4, MCNP6, and PHITS (Particle and Heavy Ion Transport code System) codes for initialization time, computation speed, and memory requirements. The computational speed results showed that the male TM phantom on the PHITS code was 2–3 times faster than the male voxel-type reference phantom on the PHITS code for all particles (e.g., photons, electrons, and helium ions) and energies considered. The TM phantom on the Geant4 code was even faster than the voxel phantom on the PHITS code for photons and electrons by up to 20 times. However, the TM phantom on the MCNP6 code was slower than the voxel phantom on the PHITS code by up to 20 times [148].
The developed adult MRCPs were recently released through ICRP Publication 145 [149]. These phantoms are the mesh counterparts of ICRP Publication 110 voxel phantoms, which overcome the dosimetric limitations from the finite voxel resolutions and the nature of the voxel format. Hence, they replaced the ICRP Publication 110 voxel phantoms and will become the next generation of ICRP adult male and female reference phantoms in mesh format. The MRCPs include all the source and target organs and tissues required for the calculation of the effective dose, including micrometer-thick regions in the alimentary and respiratory tract organs, skin, eyes, and urinary bladder, significantly improving the accuracy of the dose calculation. These micron-scale structures could not be defined in the ICRP Publication 110 voxel phantoms at the given millimeter-scale voxel resolutions. Another notable benefit of MRCPs is that they are highly deformable and thus can be used to create phantoms of various body sizes and postures, expanding their capabilities for individualized dose reconstructions. In addition, MRCPs can be directly implemented into Monte Carlo codes, fully maintaining the accuracy of the high-fidelity mesh geometry.
Taking advantage of the highly deformable MPCPs, Lee et al. [150] developed a total of 18 percentile-specific adult male and female phantoms by modifying the MRCPs. These phantoms represent the 10th, 50th, and 90th percentile standing heights and body weights in male and female Caucasian populations. Similarly, Choi et al. [151] established a body-size-dependent phantom library by modifying MRCPs. An in-house program for automatic phantom adjustment was developed and applied for the practical construction of such a large number of phantoms in the library with minimized human intervention. The established library includes 108 adult male and 104 adult female phantoms with different standing heights and body weights, covering most body sizes representative of Caucasian and Asian populations. Furthermore, Yeom et al. [152] deformed the MRCPs into five non-standing postures (i.e., walking, sitting, bending, kneeling, and squatting) by developing and using a systematic posture change method based on an as-rigid-as-possible (ARAP) shape-deformation algorithm and motion capture technology. The organ/tissue masses of the phantoms are consistent with those of the MRCPs. These phantoms, which are based on the ICRP adult MRCPs, retain all the advantages of the ICRP adult MRCPs, such as the definition of the micron-scale regions and the accuracy of the dose calculation. Recently, following the completion of adult MRCPs, Task Group 103 also completed the development of pediatric MRCPs, which have the same advantages as those presented in adult MRCPs. These phantoms are the next generation of ICRP pediatric (newborn, 1-, 5-, 10-, and 15-year-old male and female) reference phantoms in a mesh format that will replace those of ICRP Publication 143 in a voxel format. Choi et al. [153] introduced developed pediatric MRCPs with a brief explanation of the development process and discussed their computational performance in the general-purpose Monte Carlo codes.
2. Autosegmentation to create person-specific phantoms using AI
Reference Man refers to the average population of a specific age and gender with loss of individual features. We must note the differences between the “population-averaged” prospective dosimetry needed for radiological protection under the current ICRP radiation protection system and the “individualized” retrospective dosimetry needed for accident dose reconstruction, medical dose tracking, or epidemiological studies [1]. Currently, radiation-related products and applications, such as CT and PET, have widely appeared in people’s lives. Obviously, these products and applications will introduce a relatively large dose error when using the population-averaged phantom. Some studies have suggested that the organ dose error for CT between person-specific phantoms and population-averaged phantoms can reach up to 40% [154, 155]. Therefore, the creation of the person-specific phantom is very important. Generally, the CT image is the best choice to create a person-specific phantom due to the rich anatomical structure feature data provided by the CT image. However, the key step is the accurate delineation of organs. In the past, the delineation of organs was usually completed manually by the clinician to achieve high contour accuracy. However, the manual delineation is labor-intensive and time-consuming, which places tremendous pressure on clinicians [156]. This also leads to the limited use of person-specific phantoms in clinics.
However, the person-specific phantom has been applied in radiotherapy for a long time due to the use of dose calculations that require high accuracy. With the widespread use of radiotherapy, the fast and accurate delineation of organs has been an urgent problem to be solved by oncologists. In the past few decades, many methods of organ autosegmentation have been proposed to provide fast and accurate delineation. In the early stages, most segmentation techniques used no or little prior knowledge, relying on the developer to encode their idea of what would provide good segmentation. These methods include intensity thresholding, region growing, and heuristic edge detection [157–159], which are referred to as low-level segmentation approaches. Subsequently, more advanced technologies have been developed. For example, region-based techniques include active contour level-sets, graph cut, and watershed algorithms [160–163], and probability-based techniques include Gaussian mixture models, clustering, k-nearest neighbor, and Bayesian classifiers [164, 165]. These techniques use a limited quantity of prior knowledge in the form of statistical information about the appearance and shape of organs. In the last two decades, a large amount of exploratory work has been invested in making better use of prior knowledge, such as multi-atlas-based segmentation, which has been one of the most effective segmentation approaches in different challenges [166–168]. However, its segmentation performance largely depends on the performance of deformable registration, which, in turn, depends on the similarity of the morphology of organs of interest between the atlas and the new image. All of these methods are insufficient for widespread clinical use due to poor segmentation quality. Fortunately, in recent years, the advent of deep learning methods involving convolutional neural networks (CNNs) has brought an unprecedented level of innovation to the field of image segmentation. Deep learning-based segmentation approaches have been the most powerful and dominant segmentation approaches [167, 169].
Deep learning is a specific part of the broader field of machine learning where algorithms can learn data representations on their own. Specifically, deep learning uses a CNN with multiple hidden layers to learn features from a dataset by modeling complex nonlinear relationships. The advancement of deep learning is attributed to big data, strong computer power (e.g., GPU applications), and efficient algorithms [170]. The state-of-the-art models in deep learning for organ segmentation are mainly variants of encoder-decoder architectures such as U-Net [171]. In the 2017 AAPM Thoracic Autosegmentation Challenge, which provided a standardized dataset and evaluation platform for testing and discussing the state-of-the-art automatic segmentation methods for radiotherapy, the team who obtained the first prize used a deep CNN, which was modified from the U-Net architecture [167]. Their approach reached and even exceeded the accuracy of clinicians for the segmentation of the lung, heart, and spinal cord [167]. In addition to CNN architectures, some techniques, such as data augmentation [172] and transfer learning [173], have also been proposed to improve the performance of organ segmentation. These advances have led to the exponential growth of publications on medical image segmentation with deep CNNs in recent years [174]. In particular, Isensee et al. [175] recently developed nnU-Net, a deep learning-based segmentation method that automatically configures itself, including preprocessing, network architecture, training, and postprocessing for any new task. Without manual intervention, nnU-Net surpasses most existing deep learning approaches, including highly specialized solutions, on 23 public datasets used in international biomedical segmentation competitions. In summary, by using deep learning, the quality of organ segmentation was greatly improved, and the segmentation accuracy can reach the level of humans for many organs.
With the development of organ autosegmentation techniques, corresponding software was also developed, such as DeepViewer (Wisdom Tech, Hefei, China; http://www.wisdom-tech.online/), which is based on the deep learning algorithm and can automatically perform the organ segmentation of CT images in a few minutes. The Dice similarity coefficient of most organs was over 0.8 [169]. Fig. 5 shows the organ segmentation results of thoracic CT images and the corresponding person-specific phantoms using this software. In addition to the automatic segmentation function, this software also has many modification functions, which is convenient for doctors to quickly make manual modifications. Currently, DeepViewer has been deployed to more than 40 hospitals in China. The development of this organ autosegmentation software will make it easier and more convenient for the creation of person-specific phantoms.

Visualization of organ autosegmentation results and person-specific phantoms based on thoracic CT images in DeepViewer software. CT, computed tomography.
In addition to radiotherapy, there have been other current studies involving the creation of person-specific phantoms. In the field of nuclear medicine, the person-specific phantom was built to accurately calculate the internal radiation dose. However, most of the current studies in nuclear medicine still use manual segmentation [176, 177]. In CT organ dose evaluation, more studies have used autosegmentation techniques to create person-specific phantoms. For example, Fu et al. [178] used a deep learning technique to contour major organs in the thorax and abdomen, and smaller organs and structures were contoured by an atlas-based technique; Lee et al. [155] used a multi-atlas label fusion technique to contour major abdominal organs in approximately 30 minutes per patient; Peng et al. [154] used a deep learning technique to contour major thoracic or abdominal organs in approximately 5 seconds per patient. These studies suggested that current autosegmentation technology greatly saves the construction time of the person-specific phantom. This makes it possible for the person-specific phantom to be widely used in clinical practice. In the future, we believe that autosegmentation technology, especially deep learning-based algorithms, will be widely used in the creation of person-specific phantoms.
3. Real-Time Monte Carlo to Calculate Organ Doses
To calculate organ doses, computational phantoms are combined with Monte Carlo radiation transport simulation methods. Monte Carlo methods can account for all aspects of particle interactions within 3D heterogeneous media, such as the human body. In a Monte Carlo code, random numbers are used to determine the distance and outcome of a particle by comparing interaction probabilities for every geometrical region of interest. This rather tedious process is repeated for an extremely large number of particles (often exceeding 1 billion), and each particle is tracked in the 3D anatomical model until all its energy is absorbed or the particle escapes from the transport geometry. The inherent statistical uncertainty can be controlled to be less than 1%, which is often more precise than an experimental result performed in a physical phantom using a dosimeter (for quantities such as the absorbed dose). With improvements in computer affordability and computing power over the last 30 years, Monte Carlo codes are essential in many applications in nuclear engineering, health physics, and medical physics. Nearly all existing Monte Carlo codes can handle CSG shapes, including voxels. There are many comprehensive reviews or introductory articles about Monte Carlo methods for health physics and medical physics [179–184]. Some of the public-domain, general-purpose Monte Carlo codes used for radiation dose calculations include EGS [185], FLUKA [186], Geant4 [187], MCNP [188], MCNPX [189], MCNP6 [190], and PENELOPE [191]. In addition, there have been specific Monte Carlo codes for radiotherapy [183]. However, there is an obvious drawback to Monte Carlo methods. The statistical nature of Monte Carlo methods requires a long computation time to reach an acceptable level of precision, and as a result, Monte Carlo codes are often not used in routine clinical applications that demand a quick response [184]. Monte Carlo algorithms are ideally suited for parallel computing, and certain Monte Carlo algorithms are considered “embarrassingly parallelizable” [188]. In fact, high-performance parallel computers or cloud computing can be used for large scientific problems. Many users, especially those in the clinical setting, still prefer desktop computers that are affordable, accessible, and private.
In the past several years, researchers have realized that GPU technologies, which were originally developed for computer games, can be used to meet the needs for low-cost, high-performance general-purpose computing. With intrinsic parallel “stream processors,” GPUs are attractive in terms of cost, space, privacy, and ease of access when compared with other technologies such as clusters, supercomputers, and cloud computing. As illustrated in Fig. 6, a GPU device offers 7.7 times more single-precision FLOPS (floating point operations per second) than the traditional CPU.
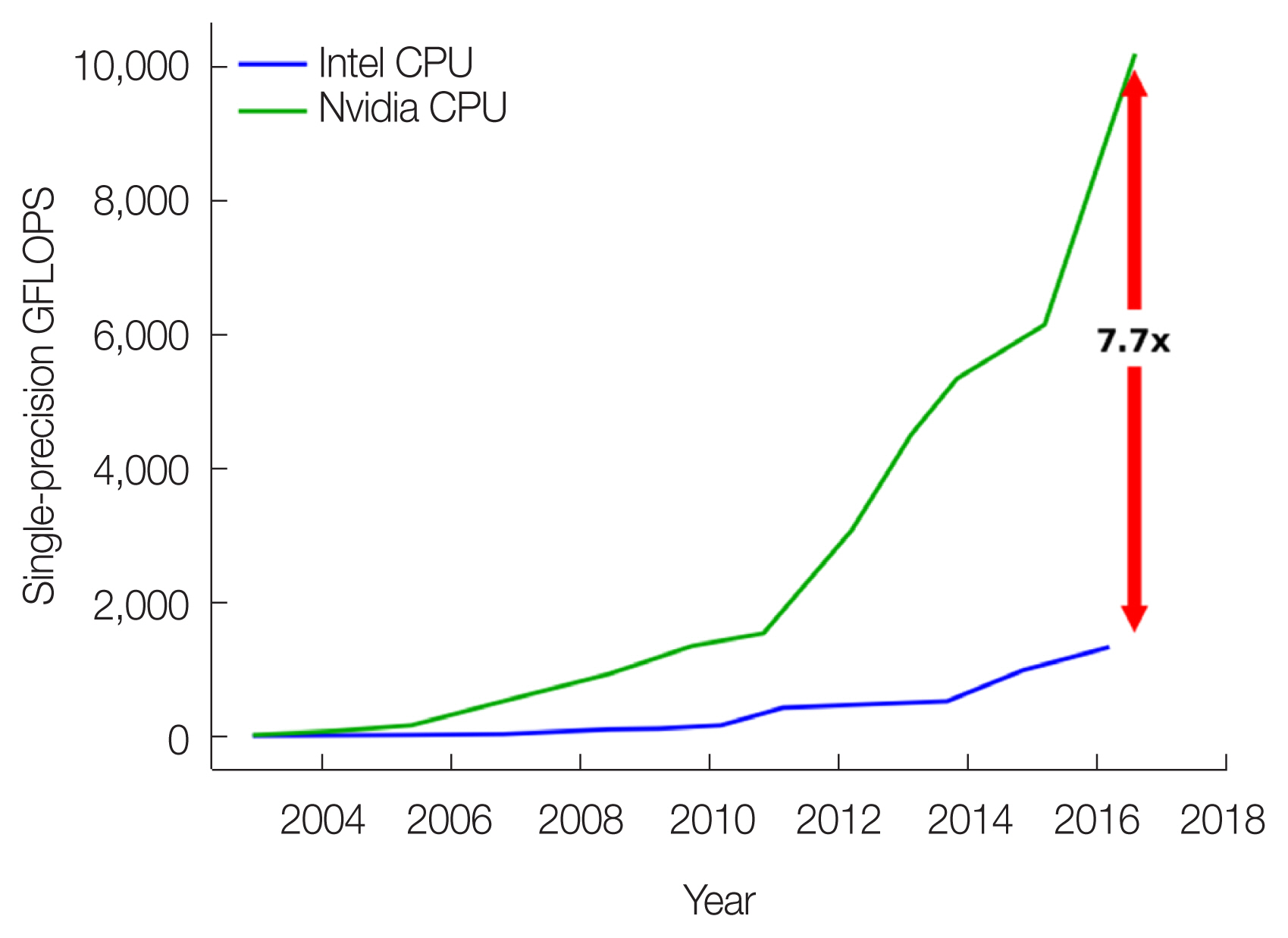
GPUs offer more computing power than CPUs. Data from NVIDIA (https://docs.nvidia.com/cuda/archive/10.0/pdf/CUDA_C_Programming_Guide.pdf). GFLOPS, giga floating point operations per second.
In the past several decades, an increasing number of top-ranked supercomputers have adopted GPUs to boost computational efficiency. GPU technologies will continue to advance rapidly as research activities in artificial intelligence, including deep learning, big data, and autonomous vehicles, have just begun to surge. However, despite exciting hardware advances, most existing production Monte Carlo radiation transport codes cannot run on GPU devices because GPUs operate on a different software operating system. For example, the software operating system for NVIDIA products is called CUDA.
As reviewed by Pratx and Xing [192] and discussed by Jia and Xu [193], the radiology/oncology community has found the power of massively parallel GPU technologies to be extremely appealing. Several recent studies have attempted to use this technology for different applications of ionizing radiation transport simulations. Badal and Badano [194] ported the general-purpose PENELOPE code into the GPU/CUDA environment for X-ray simulations to achieve a 27-fold increase in speed. Jia et al. [195, 196] developed a GPU-based Monte Carlo coupled electron-photon transport code, gDPM, for radiation treatment dose calculations. Their speedup experiment involved water-lung-water/water-bone-water phantoms and 20-MeV electron point source/6-MV photon point sources. However, only a six-fold increase in speed was observed because there was no attempt to optimize the code for GPU/CUDA. To plan radiation treatment, Hissoiny et al. [197] developed a detailed GPU/CUDA electron transport code called GPUMC. A simplified photon-electron coupled transport model and a new GPU implementation algorithm were adopted, and a 200-fold increase in speed over the DPM code was achieved. Recently, Jia et al. [195] adopted algorithms by Hissoiny et al. [197] and achieved a 69- to 87-fold increase in speed. Tickner [198] offered some insights into why most existing efforts have not yielded high expectations and proposed a number of innovative approaches associated with system of multiple initial points (SMIP) architecture, clock speed, and complex memory allocation schemes. His photon Monte Carlo code for the energy range of 1 keV–100 MeV yielded a 35-fold increase in speed over the general-purpose EGSnrc code. Jia et al. [199] also reported a fast Monte Carlo code gCTD for person-specific CT/CBCT dose calculation. A 400-fold increase in speed for the homogeneous water phantom and a 76.6-fold increase in speed for the voxel phantom compared to the general-purpose EGSnrc code was reported. Work at RPI has led to the development of a GPU-based Monte Carlo code, Accelerated Radiation-transport Computations in Heterogeneous EnviRonments (ARCHER) [200], as a testbed for diverse applications involving nuclear reactor analysis, CT X-ray imaging dosage [200], and radiation therapy [201]. As detailed in two Ph.D. dissertations, this group took a very unique approach compared to other groups [202, 203].
The Monte Carlo method is taken to be the gold standard for a large body of research work found in nuclear engineering, health physics, and medical physics. Therefore, recent attention to circumvent the bottleneck of time-consuming Monte Carlo calculations using GPUs is justified, and the approaches in leveraging extremely successful gaming technologies appear to be innovative. Many early studies by researchers in the Monte Carlo community showed the feasibility of unprecedented subminute or subsecond Monte Carlo dose calculations. However, early adopters of GPU technology were criticized for their often-biased comparison of performance against CPUs. It has also been noted that many parallelization schemes for Monte Carlo acceleration, such as vectorized methods, are not truly new. Whether GPU-based Monte Carlo calculations are just hype and how much we should invest in this relatively new technological development are issues that remain to be addressed today [193].
4. Examples of person-specific phantoms and GPU-based Monte Carlo methods
To illustrate how computational phantoms and Monte Carlo methods have advanced, projects performed by students at RPI and the University of Science and Technology of China (USTC) are described in the examples below. The topics of these projects cover health physics, diagnostic imaging, and radiotherapy.
1) GPU-based ARCHER Monte Carlo code for CT imaging and radiation treatment dose calculations
Starting in 2009, Liu and Su became engaged in an ambitious project to develop a new Monte Carlo code, named ARCHER, which is a software testbed designed to support research on Monte Carlo methods with different hardware platforms, including multiple-core CPUs, NVIDIA GPUs and Intel Xeon Phi coprocessors [146, 200, 202]. The hardware has evolved continuously in the past several years. The early system we developed for the ARCHER project used a traditional CPU as the host and multiple GPUs by NVIDIA as the “devices,” forming a “heterogeneous computer architecture.” We also considered a competing technology called the “many integrated core (MIC)” by Intel. To reduce commercialization risks, we designed ARCHER to be compatible with both NVIDIA and Intel technologies. We eventually learned that the task of developing an optimized Monte Carlo code for the CPU-GPU or CPU-MIC system was extremely challenging.
The ARCHER code was first developed for CPUs using a single thread for comparison purposes before it was revised for the GPU platform to evaluate the code performance under two different computing environments [200, 202]. Statistical errors were kept at 1%. The hardware devices included NVIDIA K20, K40, and M2090 GPUs and an Intel Xeon Phi 5110p coprocessor. The GPU/CUDA system supports the full use of the C++ programming language. It contains a new error-correcting code (ECC) memory technique to ensure large-scale computing accuracy. The hardware features a memory bandwidth of 3,144 GB/s per GPU and delivers up to 515 giga floating point operations per second (GFLOPs) of double-precision peak performance. The results for two medical physics applications are presented below: (1) X-ray CT dose calculations and (2) external beam radiation therapy dose verification.
Liu [200] also led the development of ARCHER for fast and accurate organ dose calculations based on CT images. The GE LightSpeed 16 CT scanner (GE Healthcare, Waukesha, WI, USA) was utilized, and a library of human phantoms was modeled. For performance comparison, the same random number generator and similar compiler options were used, and the hardware models were configured into the server mode. The organ dose results and computation time were benchmarked against the Monte Carlo code MCNPX. For a whole-body scan simulation, the organ doses by ARCHER were found to be in good agreement with those by MCNPX, with an average difference of 0.26%. This result suggests that ARCHER accurately simulated photon interactions [146, 200]. Since we designed all CPU, GPU, and MIC devices to run the same ARCHER code, we were able to focus on only the device performance, excluding potential software engineering effects. The computation times showed that GPU and MIC devices were able to achieve a 38- to 100-fold increase in speed compared with the CPU running on the same ARCHER radiation transport algorithms. It was noted that it took the MCNPX code 476 minutes to achieve the same precision using a CPU server of 12 message passing interface (MPI) processors. The data presented here demonstrates that the GPU- and MIC-based ARCHER codes provide an extremely cost-effective way of accelerating Monte Carlo dose calculations. Since a typical CT scan only needs to cover a portion of the body and we can use as many as six GPU cards in our system, a common dose calculation based on CT images and ARCHER can take less than a second, which is therefore considered to be in near “real-time,” when using the latest NVIDIA GPU technologies.
Su et al. [201, 202] also extended the radiological physics of the ARCHER code to include a simple version of electron transport (up to 20 MeV). This code was then used to compare with methods currently being used at the University of Wisconsin Medical Center for head, neck, lung, and prostate cancer treatment cases. A radiation treatment machine known as the “TomoTerapy electron accelerator” was simulated using ARCHER. Patient CT images were converted to voxel phantoms for dose calculations. The dose distribution of accelerator phase-space files incident on a homogeneous water phantom was used in the comparison against those obtained clinically using the production Monte Carlo code Geant4.
2) Method of virtual source modeling for external photon radiotherapy and its clinical application in dose checking
Xu [203] developed a virtual source model for external photon beam radiotherapy and integrated it into the dose verification software ArcherQA. When using the Varian TrueBeam or Elekta Axesse, the the depth doses by a virtual source model depth doses agree within 2% with the measurements, and the lateral doses are both within 3%. The dose profiles of ARCHER and EGSnrc are consistent with the film measurements, while the profile of Elekta Monaco deviates significantly from the film measurements. For clinical cases, ARCHER.QA and Eclipse have a better degree of compliance. The 3 mm/3% and 2 mm/2% gamma passing rates are above 96% and 90%, respectively. This work has resulted in a breakthrough for Monte Carlo independent dose validation software in China.
3) Organ segmentation using machine learning and person-specific CT organ dosimetry
Peng et al. [154] developed an automatic multiorgan segmentation method using a CNN model that was trained with two publicly available CT databases involving a total of 103 patients. This method performs automatic multiorgan segmentation for one patient in less than 5 seconds and has achieved good segmentation accuracy for the purpose of CT organ dosimetry in the testing cases considered in this study. The organ dose calculation method is completed in less than 4 seconds and requires 1×108 photons using the GPU-based rapid Monte Carlo code ARCHER to achieve an organ dose statistical uncertainty better than 0.5%. These results demonstrate, for the first time, the excellent accuracy and efficiency of a streamlined person-specific organ dosimetry computational tool. Implementation of such methods as part of the clinical workflow can yield considerable improvements over the current CT organ dose methods that are based on population-average phantoms, thus opening the door to prospective person-specific optimization features in the future.
4) Whole-body organ dose database for radiotherapy based on autosegmentation and fast Monte Carlo calculation
Qiu led a team to develop a whole-body organ dose database for radiotherapy based on autosegmentation and a fast Monte Carlo calculation. The autosegmentation software DeepViewer [154, 169], which is based on deep learning, was utilized to obtain organ segmentation information on systemic CT scans of a case of a patient with esophageal cancer. Then, the whole-body dose distribution was calculated by using the GPU-accelerated Monte Carlo software ARCHER [146, 200, 202]. The absorbed dose of each organ was calculated by organ mapping information and whole-body dose distribution. With patient information and organ dose data, a whole-body organ dose database was constructed. Finally, the Lyman-Kutcher-Burman (LKB) model was used to expand the organ risk assessment.
5. Limitations
The organs in stylized phantoms are represented using simple geometry due to poor computer power at the time of their inception. This obvious shortcoming leads to inaccurate dosimetric results. For voxel phantoms, due to the finite voxel resolution, it is difficult to model very thin structures such as micron-scale radiosensitive tissue layers of the skin or the alimentary and respiratory tract organs. In addition, voxel phantoms are rigid, making it difficult to adjust the posture of a phantom or use a different body size. For early BREP phantoms, such as NURBS phantoms and polygon surface phantoms, a critical technical issue results from compatibility issues with the existing Monte Carlo radiation transport codes. One limitation of tetrahedral mesh phantoms is that they cannot be used to evaluate the dose of a specific person when the required accuracy of dose calculations is very high, for example, in radiotherapy. The limitation of the person-specific phantom is that it is affected by the accuracy of organ autosegmentation.
In AI-based organ autosegmentation, first, high-quality segmented datasets are required to train a good model. Currently, the segmentation accuracy for most organs in CT images can achieve the clinical standard. However, for some organs, such as the optic chiasm, AI-based organ autosegmentation needs to be improved further. This problem could be addressed by combining CT and other imaging modalities. In addition, the generalization of the AI-based model is a noteworthy problem. Organ appearance could be affected by all types of diseases or variations in image acquisition protocols, which could potentially affect the performance of an AI-based model. Fortunately, this limitation could be addressed by using datasets from multiple institutes and data augmentation techniques. To ensure the safety of the patient, a manual check by the clinician may be necessary when AI-based autosegmentation software is used. A controversial limitation of AI-based algorithms is that they provide little interpretability for understanding how and which features affect the trained network during segmentation prediction. This hinders the ability to fully understand and identify the cause behind inaccurate segmentations [204].
There are many Monte Carlo codes in the radiation protection community. However, open-sourced GPU-based Monte Carlo codes that are well-used are difficult to find. A considerable amount of work is required to convert CPU codes to GPU codes. In addition, existing Monte Carlo codes are unable to handle a “moving” target such as the dynamic heart or lung [1].
Conclusion
Xu [1] reviewed several hundreds of computational phantoms according to three types of geometric modeling techniques. This review gave rise to the surprising discovery that the number of computational phantoms followed a pattern of exponential growth in the past 50 years. This finding suggests that we were unprepared for such rapid growth in computational phantom research in recent decades, which is reflected well in a phenomenon observed by Kurzweil [205] in his best seller, “The Singularity Is Near: When Humans Transcend Biology”; human beings tend to underestimate the technological growth rate.
The history of computational phantom development has also shown that it is the need for solving practical problems, not the need for policy-making, that has determined the course of technological advancement. The need for simulating organ motions for cardiac imaging, for example, resulted in the development of the MCAT phantom by Segars et al. [103] using quadric and the super quadric surface equations and NCAT models by Segars [101] using the NURBS technology. Shi and Xu [73] adopted “the geometry-based” respiration algorithm in the NCAT phantom for radiation treatment. Later, Eom et al. [206] developed “physics-based” respiration-simulating 4D phantoms to understand and “predict” the effects of respiration on radiation treatment. Using the same approach, Lee et al. [119] developed size-adjustable pediatric models. The BREP-based pregnant females developed by Xu et al. [113] and those by Stabin et al. [134] are also examples of application-driven research that will likely continue to dominate the research horizon in the future.
In the deep learning era, AI-based autosegmentation technologies have enabled fast and accurate delineation of organs compared with manual processes. This will greatly promote the creation of person-specific phantoms in the future. The BREP-based phantoms will always be a continued research topic in the future. NURBS geometries are flexible and computationally efficient, but fine details may be lost on certain organs that have complex topologies. Polygonal mesh geometries can be used to create very smooth surfaces with an impressive amount of anatomical detail at the cost of an excessive number of vertices. However, the calculation speed of the Monte Carlo simulations is slow. Tetrahedral mesh geometries greatly improve the calculation speed of the Monte Carlo simulations for BREP-based phantoms. Geometrical modeling of the human body is a challenge because it consists of organ surfaces of complex and unique shapes. For cardiac and respiratory motions in the frequency range of 10–100 cycles per second, mesh models may still be appropriate. However, previous work has also shown that NURBS primitives are easy to adopt for both real-time and non-real-time applications. Therefore, specific strategies will likely be based on the applications and user preferences. Regardless of the specific BREP data structure, there is currently an urgent need for application-based software that can streamline the segmentation process.
AI or deep learning provides powerful weapons for work in “modeling of digital human,” in terms of a computational phantom [43]. Automatic multi-organ segmentation is a direct beneficiary of the latest breakthrough. With new hardware technologies such as GPUs and MIC coprocessors, we have demonstrated the feasibility of near “real-time” Monte Carlo dose calculations involving a voxel and BREP phantom [200, 202]. “Real-time” MC simulations—a computational task accomplished in less than one second—is a visionary concept that is being demonstrated today [193]. In the past several years, the timing of GPU-based Monte Carlo simulations has been steadily decreasing, from 1,000 to 100 seconds, and has now decreased to a few seconds. Therefore, it is reasonable to expect that “real-time” Monte Carlo simulations will soon become a commonplace. Such a Monte Carlo simulation capability, associated with the autosegmentation of tomographic images, will likely further increase the rate of computational phantom research toward the 4th generation of “person-specific” and “multiscale” phantoms. Such phantoms will contain deformable anatomies that are physics-based and are, therefore, biomechanically realistic in depicting real-time and multiorgan deformation associated with cardiac and respiratory motions. These phantoms will also possess physiological and functional information of the human body at the organ and cellular levels obtained from emerging radiology devices [43]. By 2030, breakthroughs in computational radiobiology, in the context of cancer radiotherapy and radiobiological effects, are expected to bring a new horizon to personalized radiation medicine by understanding and harnessing the massive power of genomic data. Real-time Monte Carlo calculations will be performed routinely in shielding design and radiation oncology clinics. As the past 60-year history has revealed to us, coordinated and cooperative efforts among radiological engineers, computer scientists, biologists, and clinicians will always be the key to the success of future research endeavors.
ACKNOWLEDGEMENTS
This work was supported in part by University of Science and Technology of China (USTC) grants on “New Medicine Team Project: The ROADMAP Medical Physics Platform” and “Med-X Medical Physics and Biomedical Engineering Interdisciplinary Subjects” Strategic Priority Research Program (No. XDB39040600), and in part by Natural Science Foundation of Anhui Province, China (No. 1908085MA27).
Notes
CONFLICT OF INTEREST
The authors of this manuscript declare relationships with the following companies: Virtual Phantoms Inc. and Wisdom Tech Inc.
AUTHOR CONTRIBUTION
Conceptualization: Xu XG. Methodology: Xu XG, Chen Z, Peng Z. Formal analysis: Peng Z. Funding acquisition: Xu XG. Project administration: Xu XG, Chen Z, Peng Z. Visualization: Gao N, Wu B. Writing - original draft: Peng Z, Gao N, Wu B. Writing - review and editing: Xu XG, Chen Z, Peng Z. Approval of final manuscript: all authors.